EXPERIMENTAL EVOLUTION OF THE COGNITIVE BRAIN

Evolution results in a marvelous diversity of phenotypes whose sole function is to support the propagation of the genetic code. What drives the ever-increasing complexity of adaptive traits has been a core focus of evolutionary biology. One of the greatest evolutionary mysteries has been the evolution of cognition. What kind of selective pressure was necessary to produce a brain capable of understanding the intricacies of the universe and the brain itself? In which environments are such extreme phenotypes adaptive? And how do we select for a trait that is inherently extremely plastic at the level of individual? These are challenging questions that have been so far tackled using comparative and correlative approaches across animal phylogeny. Our lab on the other hand, aims to advance the field by moving forward from indirect inference of cognition in natural populations to direct experimental evolution of brain's structure and function in the lab. The major goal of our group is to push the limits of cognitive neuroscience by using evolution as a tool to study the mechanistic basis of cognitive ability. We leverage the genetic, phenotypic and experimental utility of the model fruit fly, Drosophila melanogaster, and the power of experimental evolution to map the genetic, structural and functional underpinnings of adaptive cognition in diverse populations. By developing high-throughput phenotyping methods that combine real-time imaging and machine learning techniques, robotics and automatization, we can select for individuals exhibiting high and low cognitive ability in whole experimental populations of Drosophila over each generation and "induce" the evolution of cognition. At the same time, we plan to also manipulate natural selective pressures that have been hypothesized to influence the evolution of the brain, such as social interaction or environmental complexity. In a way, instead of reverse engineering already cognitive brains, we forward engineer cognition experimentally in a cognitively modest animal system. Moving the study of evolution of brain from past selection to real-time, present, and controlled selection will bring us closer to directly answering a long-lasting mystery: how does a brain evolve and give rise to cognition?
NATURAL GENETIC VARIATION OF COGNITION

Genetic variation is the essential substrate for evolution. To understand how and why some variant are selected while others are not, we first need to understand the breadth of variation that is available to selection. Our lab aims to identify natural phenotypic variation underlying neuronal traits such as neuronal and brain morphology, biological processes involved with cognition such as learning and memory, decision making and perception, and variation in neuronal structure underlying cognition. Using the extensive genetic resources available for Drosophila melanogaster and developing new imaging techniques we aim to map the natural genetic variation underpinning the connection between brain structure and brain function. Understanding natural genetic variation of neuronal traits will help us better understand their mechanistic basis from genetic, to molecular, structural and behavioral level. While we used to think that cognition could be studied at the level of genotype within a single generation, our recent findings have steered our research to the level of individual. We believe now that the genetic variation of cognitive ability is best approached transgenerationally, while plastic molecular and structural variation of the brain may point to variation in cognitive ability across individuals, within a generation.
ROBOTIC AUTOMATION OF DROSOPHILA GENETICS AND BEHAVIOURAL PHENOTYPING
Whether its goal was to identify a rare mutation, map a genetic locus, or characterise a gene's function, the field of genetics has always relied on sampling probabilities. Hence, its success has and still does rely on high throughput phenotyping and genotyping of large sample sizes. Throughout the history geneticists have been using established manual manipulation and screening techniques to perform the aforementioned tasks. But the pace of today's scientific progress has made manual screening techniques we have been using since 1900' entirely obsolete. The exceptional demand of our lab's main research projects for wast amounts of phenotyping and genotyping data and our innate drive to employ cutting-edge technologies of the 21st century in our research pipelines created a perfect storm for the development of fully automated, robot-assisted fly genetics platform we call "Robogenetics". Our aim is to fully automate fly rearing, handling, phenotyping and genotyping using a collaborative robotics platform and machine-vision accessible genetic markers. The goal of this project is to design and create a plug-and-play system that is technologically as well as financially accessible to an average fly lab which will enable hands-off high-throughput Drosophila genetics experiments, especially those aimed to screen and investigate the genetic basis of behavioural traits, or more translational applications such as automated compound, drug and genetic screening for neurological disorders with behavioral symptomatology.
COLLABORATIONS
WITH GRÄFF LAB (EPFL SV GRÄFF LAB)
Cognitive fitness from a genetic, metabolic, and evolutionary perspective
The risk for developing Alzheimer’s disease (AD) is inversely correlated with the degree of educational attainment: The higher the educational achievement, the lower your risk for AD. This association is called the “cognitive reserve hypothesis” and, although epidemiologically supported by multiple studies, the physiological and molecular mechanisms behind this association are poorly understood. In this project, we set out to test the cognitive reserve hypothesis from an energy demand perspective. Since both learning and neurotoxicity in AD pose a high demand on cellular energy consumption, we hypothesize that well-trained brains are conditioned to utilize energy more economically. In other words, energy saved by a well-trained brain on learning can be re-routed to protect against neuronal cell death. To experimentally assess this hypothesis, we will use D. melanogaster we well as murine models of AD in a joint project between the Jakšić and Gräff labs. Specifically, we will first assess the genetic basis for the relationship between lifetime learning, neurodegeneration, energy consumption, learning-dependent metabolic rates in a screen of genetically characterized D. melanogaster lines. These results will then be validated in different mouse models of AD. Knowing the genetic architecture underlying metabolism, lifetime learning and the risk for AD will yield important insights into molecular basis of the cognitive reserve hypothesis.
WITH REIS LAB (EPFL IGM FLEX LAB)
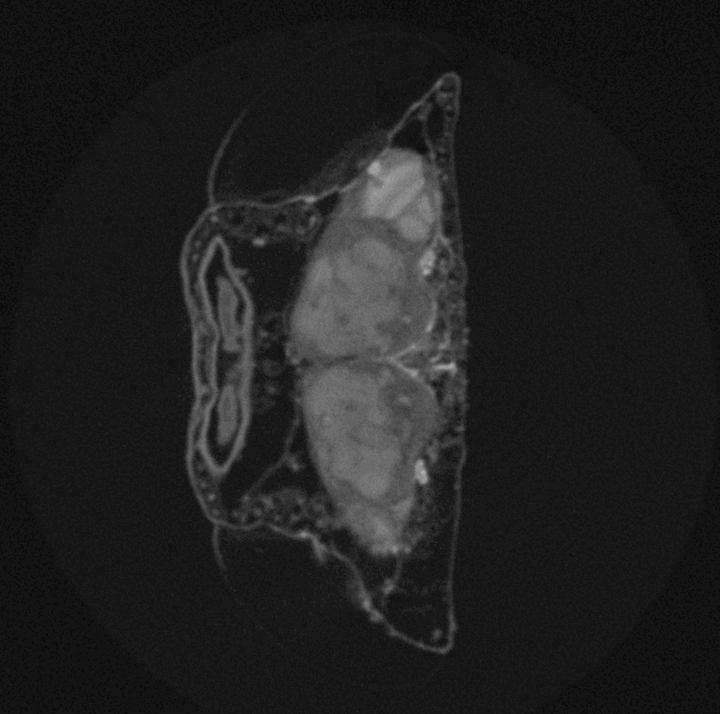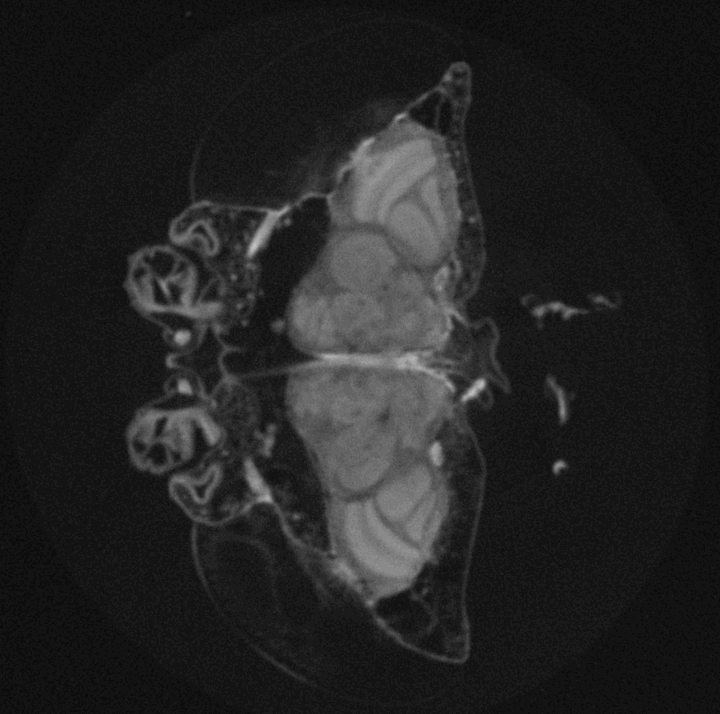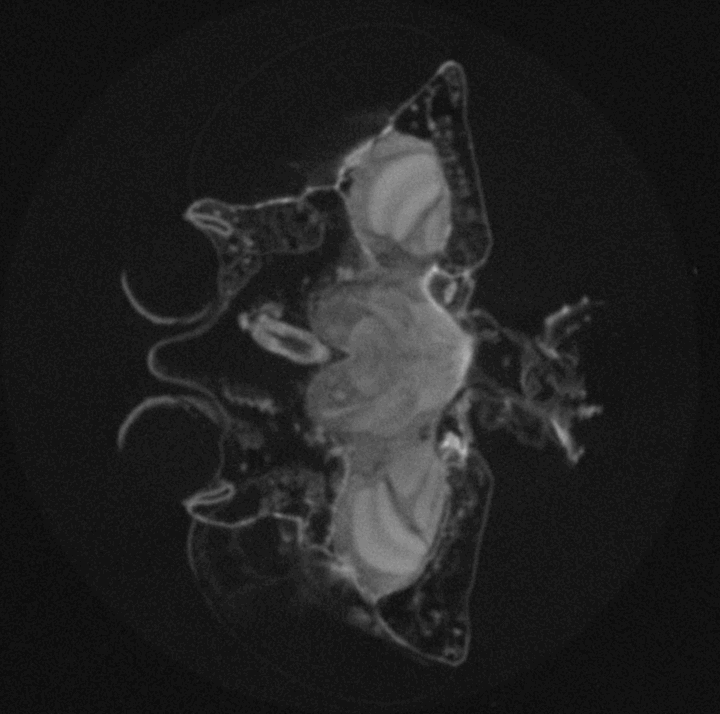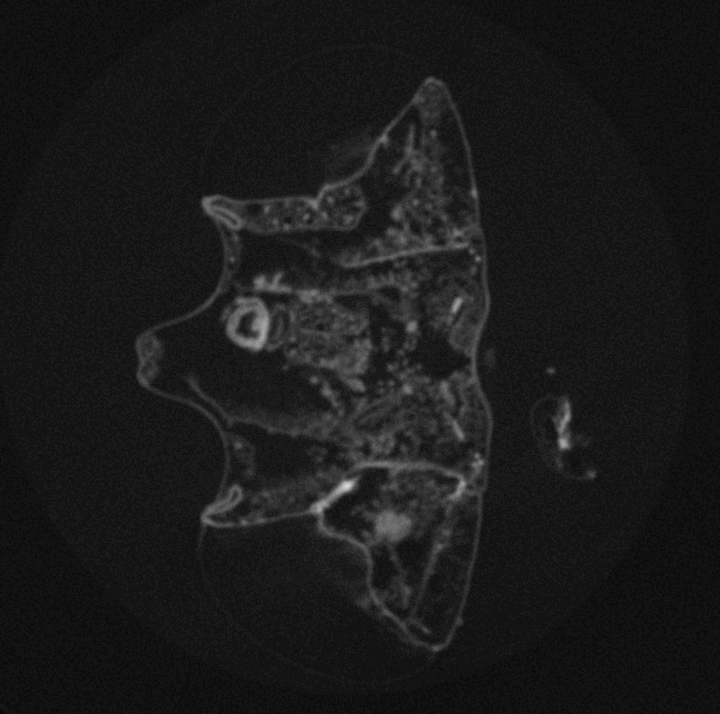
High-throughput Drosophila brain morphometrics using nano- and µ-Computed Tomography (µCT) and Topological Data Analysis
Around 2 million years ago brain size of the hominid lineage started growing, and quickly doubled in size. This tremendous increase has served as a morphological proxy for evolution of extreme cognitive ability we observe in modern human populations. While skeletal remains can reveal brain size of ancient populations via measurements of intracranial volumes, any behaviours that may have emerged from brain size increase is lost to the past. However, experimental evolution of cognition provides a window into the connection between evolution of behaviour and evolution of brain morphology as both can be measured in real-time, as they evolve. The challenge is to reliably measure fine, naturally occurring variation in whole-brain morphology in whole populations and across scales. In this project we combine the expertise of our two labs to develop a method to rapidly measure brain morphology of naturally genetically diverse fly populations. This method will help us track the evolution of brain size in real-time, as well as identify genetic mechanisms underlying brain morphology and its correlated behavioural outcomes. We also find its use in structural characterization of other biological tissues, beyond the Drosophila brain structure.
Link to abstract (Neurofly 2022)
Samuel Bourgeat (PhD student Jakšić Lab)
Fani Derveni (Postdoc, Reis Lab)
WITH JOOST LAB (EPFL ENAC LASIG LAB)
Molecular adaptation of corals to heat through the Coral Sea (MACH)
If not prevented, rapid coral reef bleaching caused by global warming will lead to total local ecosystem collapse. Fortunately, it has been observed that some coral populations exhibit heat-stress resistance but the underlying cause is unknown. In this study we combine environmental, genomic and physiological data along a the Coral Sea's reef cline to create the most comprehensive spatial catalogue of heat-adaptive genetic repertoire of a cornerstone reef building coral species Acropora millepora. The ultimate goal is to use this knowledge to guide concrete reef conservation efforts by taking into account genetic strengths and weaknesses of threatened coral species.
Joint PhD student Manon Pribille
Funding: iPhD program (2021 call)


